Rapid determination of stability by monitoring aggregation and degradation in a single measurement.
Speed, accuracy, and simplicity
Mass photometry is an innovative method for analyzing molecules. It enables the accurate mass measurements of single molecules in solution, in their native state and without the need for labels. For high-sensitivity, rapid, and user-friendly molecular mass measurements, a TwoMP mass photometer offers wide mass range and single-molecule resolution.
Key Features
1. Accurate measurements of molecular mass
2. Minimal quantities of sample required
3. Intuitive acquisition and data analysis software
4. Easy setup – a compact, benchtop instrument with minimum installation requirements
Applications
1. Protein-protein interactions
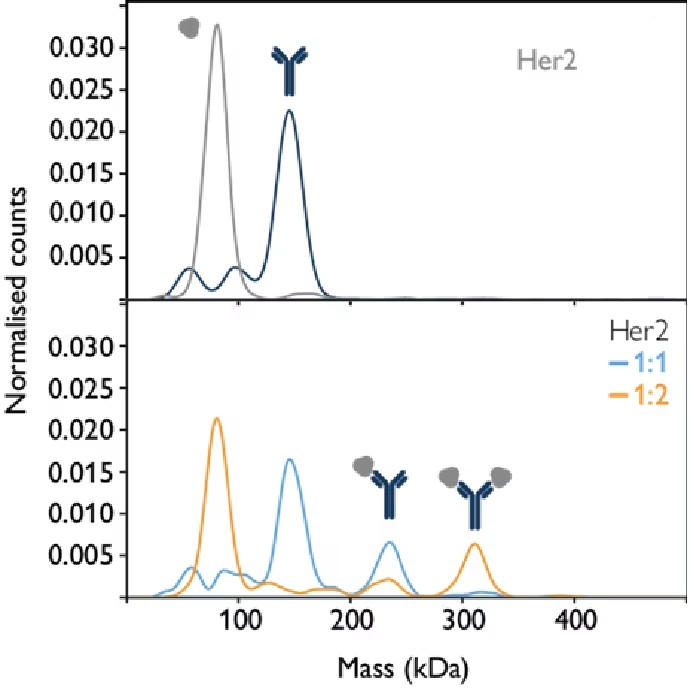
Antigen-antibody interactions are a prime example of molecular systems that can be studied using mass photometry, which can be applied to determine binding affinities for mono- and multivalent interactions.Figure 1 illustrates how mass photometry can be used to quantify molecular interactions. By measuring the mass of an antibody and its target antigen, Her2, individually and in mixtures, the interactions between individual antibody molecules and target antigens could be quantified.
2. DNA-protein interactions
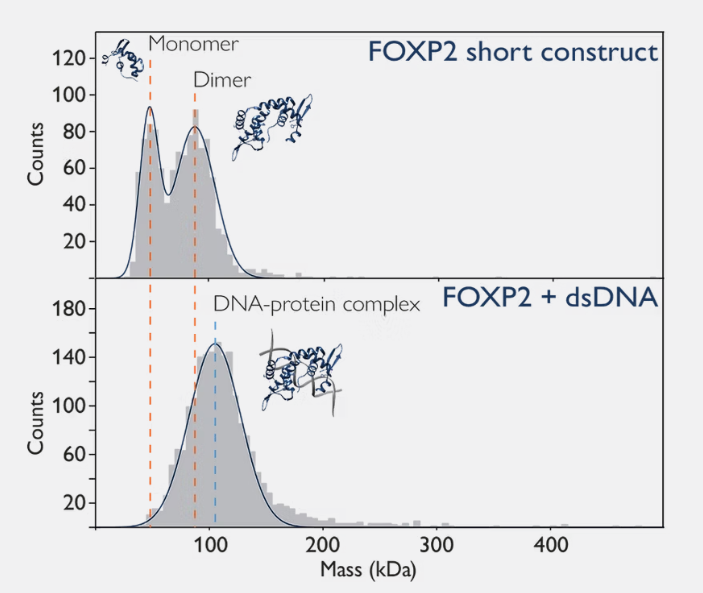
Mass photometry can be used to characterise DNA-protein complexes, which are crucial in gene expression, replication and DNA repair.In this example, mass photometry not only allows detection of DNA binding but also provides information on how the oligomeric state of the protein changes upon DNA binding (Figure 2).
3. Protein oligomerization
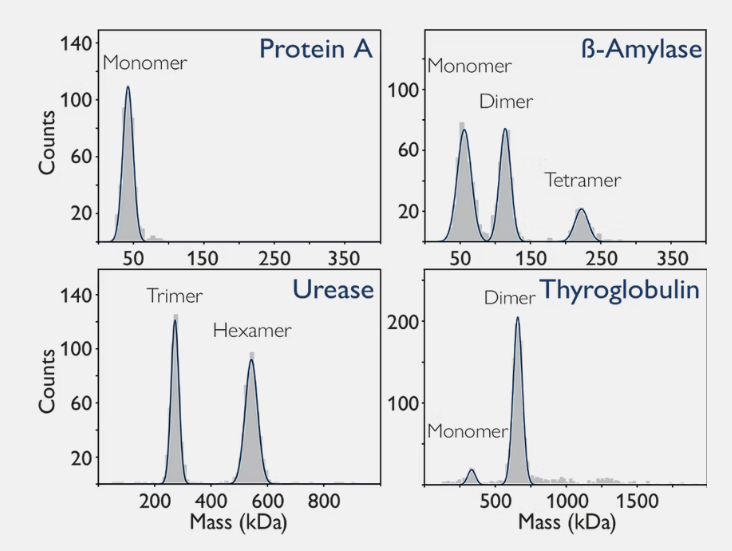
Many proteins adopt a particular oligomeric form under certain conditions. In Figure , mass photometry characterises four different proteins: protein A, beta-amylase, urease and thyroglobulin. Through this characterisation, these proteins showcase a range of behaviours – from purely monomeric (as in Protein A) to a dynamic equilibrium between multiple states.
4. Complex formation
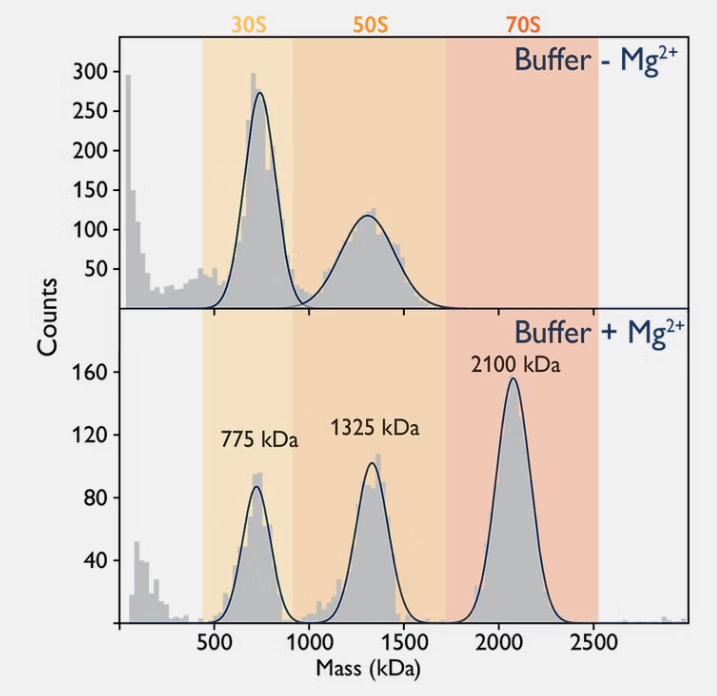
Ribosomes are macromolecular assemblies of protein and RNA, and are central sites for protein synthesis. Bacterial ribosomes (in this example from E. coli) are >2 MDa in size. Magnesium ions play a subtle yet important role in the assembly of intact (70S) complexes. In the absence of magnesium, E. coli ribosomes rapidly disassemble into their 30S and 50S subunits. Mass photometry allows us to monitor these processes (Figure).
(Please refer to the product mannual for information details.)




